5' Research Brief - Structuring Radiology Reports: Challenging LLMs with Lightweight Models
5 Minutes to get the key ideas.
Original Article: “Structuring Radiology Reports: Challenging LLMs with Lightweight Models”
Authors: Johannes Moll, Louisa Fay, Asfandyar Azhar et al
Source: ArXiv (Link)
Introduction
Radiology reports are critical for clinical decision-making but often lack standardized formats, creating challenges for both human interpretation and machine learning applications. While large language models (LLMs) show promise in reformatting clinical text, their high computational requirements, lack of transparency, and data privacy concerns hinder practical deployment in healthcare settings.
Objective
This study evaluates whether lightweight, task-specific models (<300M parameters) can effectively compete with much larger LLMs (1B-70B parameters) for structuring free-text chest X-ray radiology reports while offering advantages in computational efficiency, deployment feasibility, and data privacy. The goal is to transform unstructured reports into standardized formats following the RSNA RadReport Template Library.
Proposed Solution
The authors developed two lightweight encoder-decoder architectures: T5-Base (223M parameters) and BERT2BERT (278M parameters), initialized with various pretrained models to investigate domain-specific pretraining benefits. They leveraged GPT-4 as a "weak annotator" to generate structured versions of 182,962 reports from MIMIC-CXR and CheXpert Plus datasets, with human expert validation on 223 reports. Models were trained to structure reports according to the "RPT144" template, organizing content into sections like Exam Type, History, Technique, Comparison, Findings (by organ systems), and Impression.
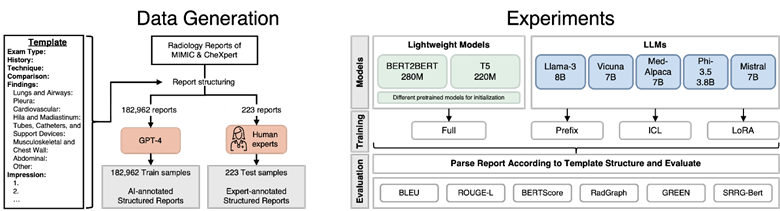
The lightweight models were benchmarked against eight open-source LLMs using three adaptation techniques: prefix prompting (instructions without weight modification), in-context learning (ICL with examples), and low-rank adaptation (LoRA finetuning modifying ~0.1% of parameters).
Evaluation Approach
Evaluation was focused on the Findings and Impression sections and employed multiple metrics on a human-annotated test set of 223 reports reviewed by five radiologists. Linguistic quality was assessed using BLEU (n-gram overlap), ROUGE-L (longest common subsequence), and BERTScore (semantic similarity). Clinical accuracy was measured through F1-RadGraph (clinical terms/relationships precision/recall), GREEN (factual correctness via finetuned LLM), and F1-SRR-BERT (disease classification into 55 labels). Resource efficiency metrics included training time, inference time, cost, and carbon emissions.
Results
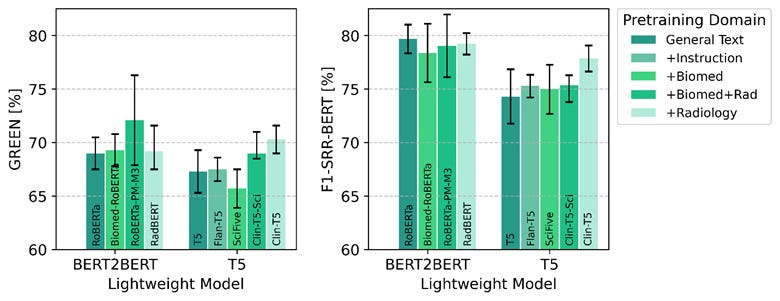
The best lightweight model (BERT2BERT initialized from RoBERTa-PM-M3) outperformed all LLMs using prompt-based techniques. Domain adaptation showed clear benefits, with biomedical and radiology-pretrained models achieving 4.5% GREEN improvement over general-text baselines.
LoRA finetuning achieved highest LLM performance, outperforming prefix prompting. Some LoRA-finetuned LLMs achieved modest gains over the best lightweight model on the Findings section but, when averaged across both sections, no LLM surpassed the lightweight model.
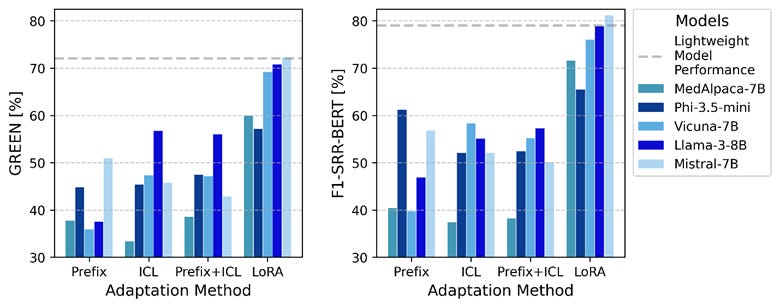
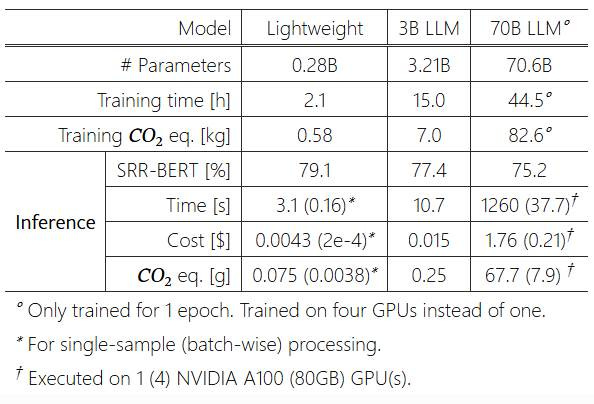
However, the computational efficiency advantages were dramatic: LLaMA-3-70B incurred over 400 times the inference time, cost, and carbon emissions of the best lightweight model
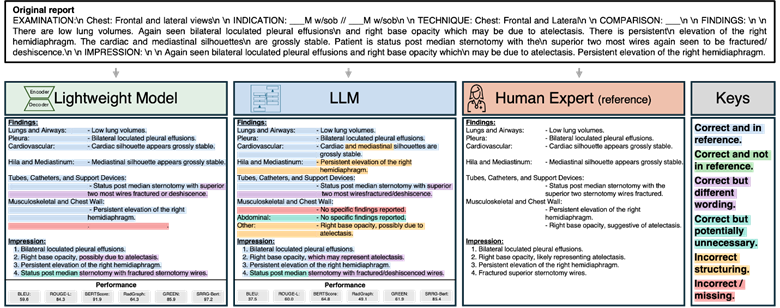
Besides, qualitative analysis showed both lightweight and LLM models correctly applied section headers in all cases, with occasional formatting variations in <1% of LLM reports. Both models included only relevant organ systems and findings, with complete omission of relevant findings occurring in <1% of cases.
Why This Matters
This research has significant implications for healthcare technology and sustainability. Lightweight models enable on-device deployment in resource-constrained settings, eliminating cloud-based solutions that raise data security and regulatory compliance concerns. Local processing addresses critical privacy concerns, aligning with HIPAA and GDPR regulations.
The dramatic computational resource reduction translates to substantial cost savings, making AI-assisted report structuring economically viable for smaller healthcare facilities. The significantly lower carbon footprint (up to 400 times less than large LLMs) represents an important step toward sustainable AI in healthcare.
This research challenges the "bigger is better" AI paradigm, demonstrating that carefully designed lightweight models can achieve comparable performance to much larger LLMs for specific clinical tasks while offering substantial advantages in efficiency, privacy, and deployment feasibility.
To Go Further
If you wish to go beyond this overview, the complete paper can be found at: https://arxiv.org/abs/2506.00200v1
Note: Unless stated otherwise, all images presented in this article are derived from the original work by Johannes Moll et al.
Key Terms Explained
Term – Definition
BERT2BERT – An encoder-decoder architecture initialized with BERT, designed for sequence-to-sequence tasks like text restructuring.
BERTScore – A metric evaluating semantic similarity between generated and reference text using contextual embeddings from the BERT model.
BLEU (Bilingual Evaluation Understudy) – A text generation metric quantifying n-gram overlap between model output and reference text.
Encoder-decoder architectures – Neural networks that compress input data into a latent representation (encoder) and reconstruct it into structured output (decoder).
F1-RadGraph – A clinical evaluation metric assessing precision/recall of extracted medical entities and their relationships in radiology reports.
F1-SRR-BERT – A metric for disease classification accuracy, scoring alignment with 55 predefined clinical labels using BERT embeddings.
GDPR– EU Legal framework ensuring data privacy
GREEN – A factual correctness metric for generated reports, computed via a fine-tuned LLM judging clinical accuracy.
HIPAA – US Legal frameworks ensuring patient data privacy
In-context learning (ICL) – A technique where models infer tasks from input examples without weight updates.
Inference time – The duration a model takes to process input and generate output post-deployment.
Large Language Models (LLMs) – AI systems trained on extensive text corpora, capable of complex language tasks but computationally intensive.
Lightweight models – Compact neural networks (<300M parameters) optimized for specific tasks, balancing performance and efficiency.
Low-rank adaptation (LoRA) – A parameter-efficient fine-tuning method modifying only 0.1% of weights via low-rank matrices.
On-device deployment – Running AI models locally (e.g., hospital servers) to avoid cloud dependencies, enhancing privacy and speed.
Prefix prompting – Guiding model outputs using instructional text prompts without altering its internal parameters.
Pretrained models – Models pre-trained on generic or domain-specific data (e.g., RoBERTa-PM-M3 for biomedicine), later fine-tuned for tasks.
ROUGE-L (Recall-Oriented Understudy for Gisting Evaluation - Longest) – A metric comparing the longest matching subsequence between generated and reference texts.
RSNA RadReport Template Library – A standardized schema for organizing radiology reports
T5-Base – A 223M-parameter encoder-decoder model variant of the Text-to-Text Transfer Transformer architecture.
Weak annotator – An AI system (e.g., GPT-4) generating approximate training labels, validated by human experts for accuracy.